Research Computing, a team within UVA Information Technology Services, recently held its second annual Research Computing Exhibition on April 23. This event showcased how researchers at UVA are pushing the boundaries of innovation using Research Computing’s services and resources.
Featuring research presentations, faculty lightning talks, and a poster competition, the exhibition underscored the growing significance and impact of computational research across the University.
More than 30 students and scholars presented posters highlighting research powered by UVA Research Computing’s resources. Their work spanned a wide range of fields — including environmental science, astronomy, cancer genomics, digital health, and chemical engineering — demonstrating the breadth of disciplines benefiting from advanced computational tools. Projects included analyzing forest dieback along the Atlantic Coast, predicting health outcomes for newborns in intensive care, and investigating the chemistry of stars.
In addition to the posters, faculty from across UVA delivered lightning talks—brief, high-level presentations on how they are integrating Research Computing tools into their research. Topics ranged from exploring the universe to advancing healthcare and molecular biology.
The poster competition was judged by a cross-disciplinary panel of UVA faculty, who evaluated submissions in two categories: Biological & Health Science and Engineering & Physical Sciences. Evaluations were based upon three criteria: 1) the extent to which Research Computing resources contributed to the research, 2) how clearly the use of those resources was presented on the poster, and 3) the clarity and significance of the scientific findings.
Below are this year’s winners of the poster competition. The winners receive a travel voucher to attend a conference of their choice where they can present their work.
Thank you to all the researchers for sharing your brilliant insights and compelling results with us!
Biological & Health Science

#1: Henry Yeung
Graduate Student, Department of Environmental Sciences
Poster: Extensive Ghost Forest Formation Across the US Atlantic Coast
Abstract:

Rising sea levels and climate-driven stressors are transforming coastal forests, reducing their ability to sequester carbon, protect against storms, and provide other services (e.g. timber) to coastal communities. Yet, the true extent and drivers on coastal forest loss remain poorly quantified. By mapping over 10 million individual dead trees across the US Atlantic region, we reveal pervasive mortality even in areas previously deemed resistant to sea-level rise –– an extent far greater than previous assessments had recognized. Previous studies only based their findings on a few well-studied sites with mass dieback, largely overlooking landscape-scale increase in tree mortality. By integrating deep learning and sub-meter imagery with the computing capacity of the Rivanna high-performance computing, we, for the first time, expose fine-scale vulnerability patterns that are invisible to traditional satellites across 1.2 million hectares of low-lying forests (< 5m), resolving untested assumptions on the roles of salinity versus flooding at regional scale. We find that these at-risk areas experience mortality fourfold higher than upland ecosystems, driven mainly by salinization. Our findings highlight an alarming yet underestimated scale of coastal ecosystem loss, offering scientists and policymakers urgently-needed insights and tangible pathways to preserve these vital habitats.

#2: Navya Annapareddy
Graduate Student, UVA School of Data Science (PhD)
Poster: A New NICU: High Performance Compute Enabled Digital Twins for Real Time Infant Monitoring
Abstract:

1 in 10 infants are born prematurely globally and receive specialized care in environments like the Neonatal Intensive Care Unit. Preterm infants are at exponentially higher risk for developmental disorders, such as cerebral palsy and autism at rates tens to hundreds of the general population. The most common diagnostic for such disorders is the general motion assessment (GMAs) heuristic carried out manually by clinicians for preterm infants to assess neurodevelopmental (NVD) risk. As formal screening and diagnosis methods are limited by availability of trained clinicians and NICU size, we propose using computer assisted digital twins for automatic, even remote, risk assessment using a computer vision machine learning (CVML) model. We successfully develop the first clinical pose estimation framework for infants in clinical care settings meant for real-time streaming contexts. Our two-step sequential framework is comprised of two distinct models: (1) a CNN model for pose estimation of anatomic key points, and (2) a digital twin model prototype to reproduce the pose estimation CNN results in high fidelity. Each frame of a video is processed by the model pipeline and is only enabled by the vastly different capabilities of the HPC ecosystem, ranging from data storage, labeling, manipulation, training, and even validation and low latency deployment onto edge devices such as clinical tablets and computer devices.

#3 Jisu Shin
Graduate Student, Biochemistry and Molecular Genetics, Biomedical Sciences Graduate Program
Poster: Multi-modal Single-cell Sequencing Reveals Cellular Heterogeneity in LGL Leukemia
Abstract:
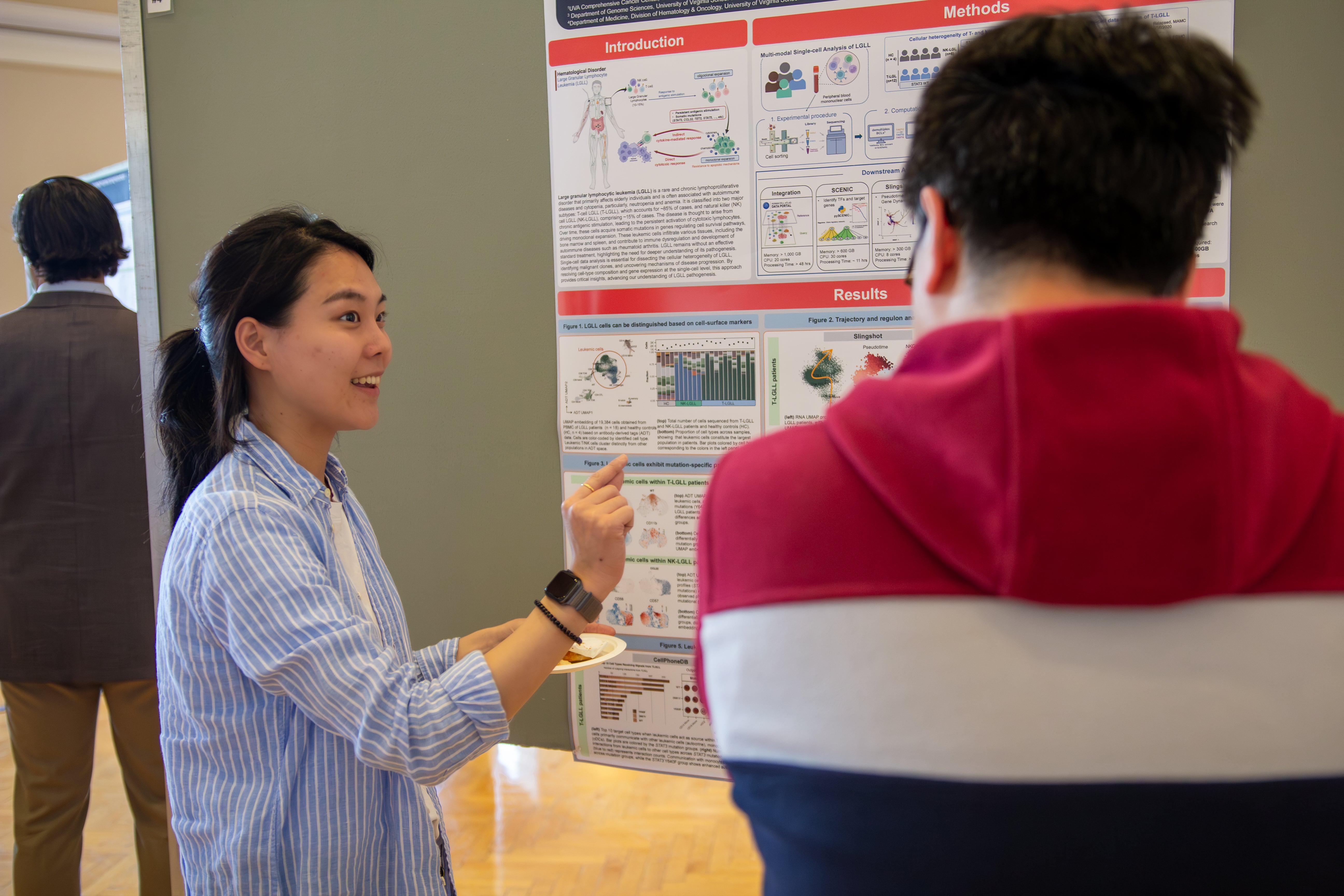
Large granular lymphocytic leukemia (LGLL) is a rare lymphoproliferative disorder characterized by clonal expansion of cytotoxic lymphocytes. To investigate the cellular heterogeneity and molecular mechanisms of LGLL, we performed single-cell multi-omics analysis, integrating transcriptomic, proteomic, and TCR sequencing data from patients treated at UVA, a national referral center for this rare disease. Given the scale and complexity of the data, our analyses required UVA’s high-performance computing (HPC) resources, Rivanna and Afton. Parallelization and batch corrections in pre-processing steps significantly accelerated data analysis, making large-scale single-cell processing feasible. However, downstream analyses required careful step-by-step validation and could not be automated through job submissions. The ability to run interactive jobs with large memory and multiple cores was essential for ensuring accuracy and efficiency in data integration and interpretation, enabling insights into LGLL pathogenesis that would be computationally infeasible on standard systems.
Engineering & Physical Sciences

#1: Mélisse Bonfand-Caldeira
Post-Doctoral Fellow, Departments of Astronomy and Chemistry
Poster: Unlocking the chemistry of stars with high-performance computing
Abstract:
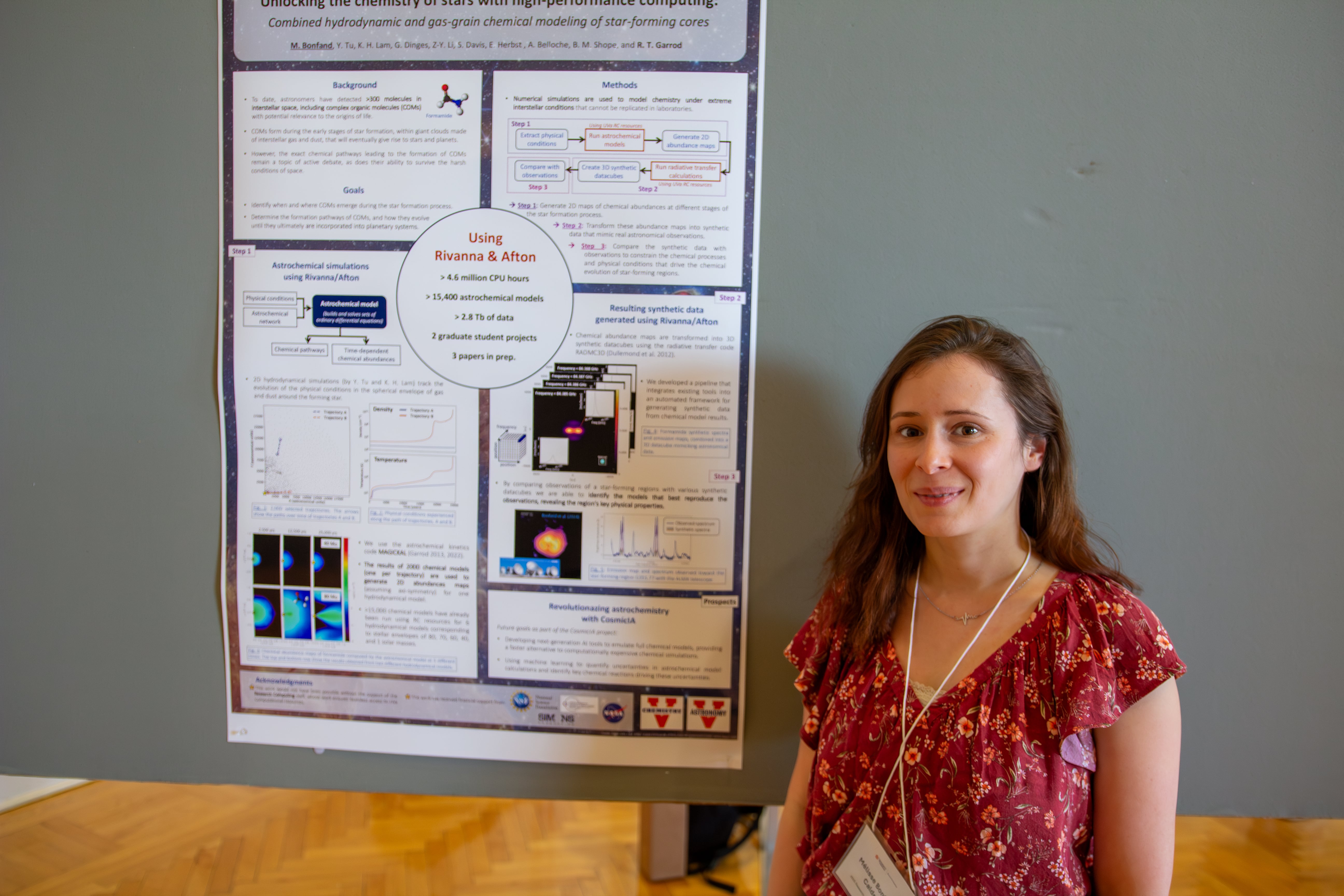
Complex organic molecules, the elemental building blocks of life, are known to form in space within giant clouds of interstellar gas and dust that eventually give rise to stars and planets. To understand how these molecules form and evolve, surviving the harsh conditions of space, until they eventually become incorporated into planetary systems, we harness the computing power of Rivanna/Afton to run thousands of numerical simulations, modeling the chemistry of various star-forming environments. We have developed an automated pipeline that then transforms astrochemical model outputs into synthetic data, mimicking real astronomical observations. By comparing observations of star-forming regions with a large grid of synthetic data, we are able to identify the models that best reproduce the observations, unveiling the region’s key physical properties and the chemical reactions that shape its molecular complexity.

#2: Md Jakir Hossen
Graduate Student, Chemical Engineering
Poster: Elucidating Biomolecular Surface Hydrophobicity
Abstract:
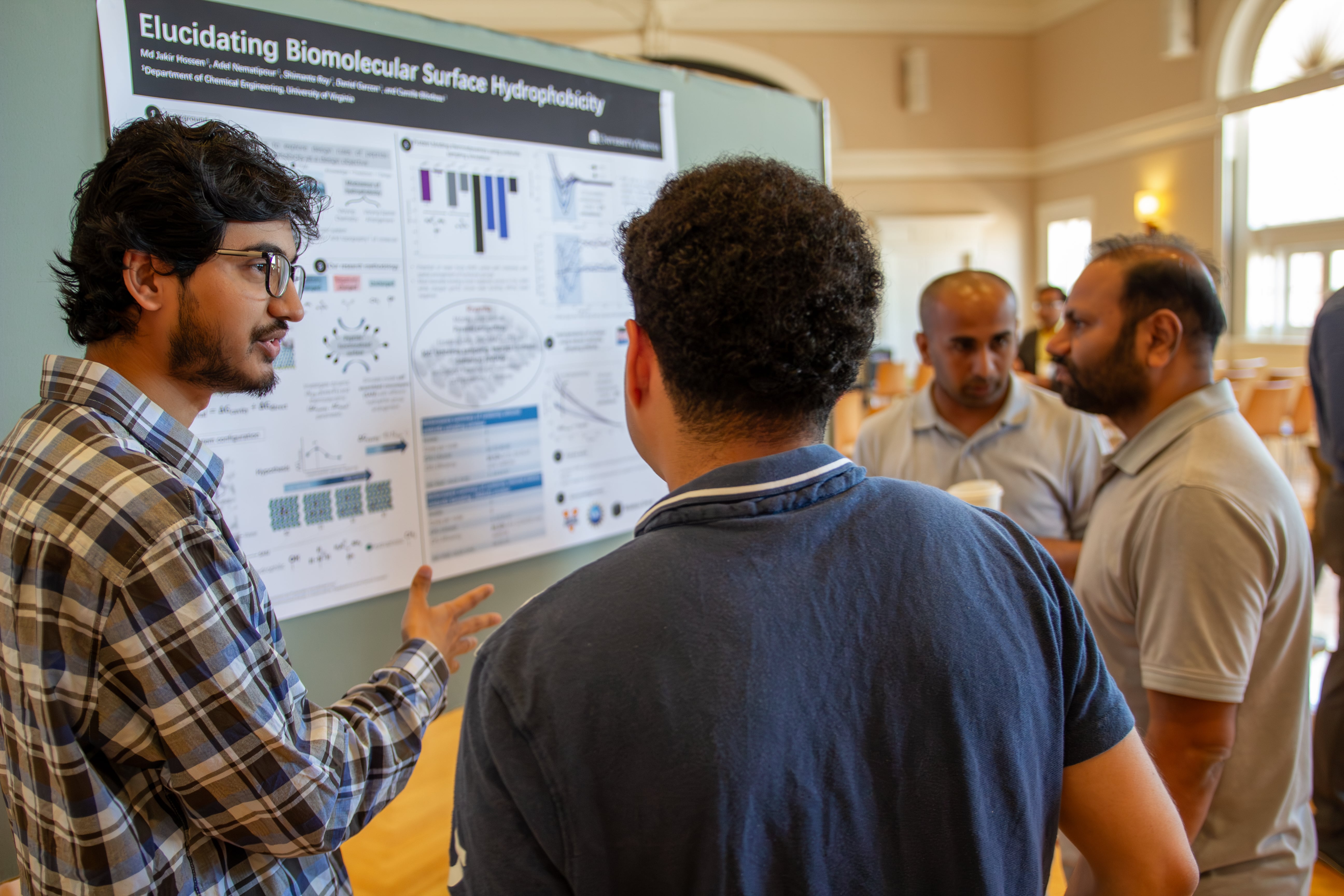
Many biophysical processes are driven by the removal of water molecules, known as hydrophobic interactions, near complex biomolecular surfaces such as proteins, enzymes, and membranes. While the hydrophobicity of individual non-polar, polar, and charged amino acids is relatively simple to predict, collective water-mediated interactions on heterogeneous surfaces remain challenging due to non-additive effects. In this study, we designed self-assembled monolayers (SAMs) with varying spacings of hydroxyl, ammonium, and guanidinium groups—common in amino acid sequences—to investigate hydrophobic behavior in a non-polar environment. Using molecular dynamics (MD) simulations on Rivanna’s high-performance computing resources, we analyzed the binding affinity of a model hydrophobic protein to these surfaces and employed indirect umbrella sampling (INDUS) simulations to compare their hydrophobicity. Our findings reveal the role of hydrogen bonding dynamics, binding affinity, and the free energy of water removal, emphasizing the impact of functional group spacing. These insights provide a foundation for studying more realistic peptide-functionalized surfaces, where such functional groups are prevalent in a non-polar background.